This talk was given at 2020-07-15 at ISMB 2020
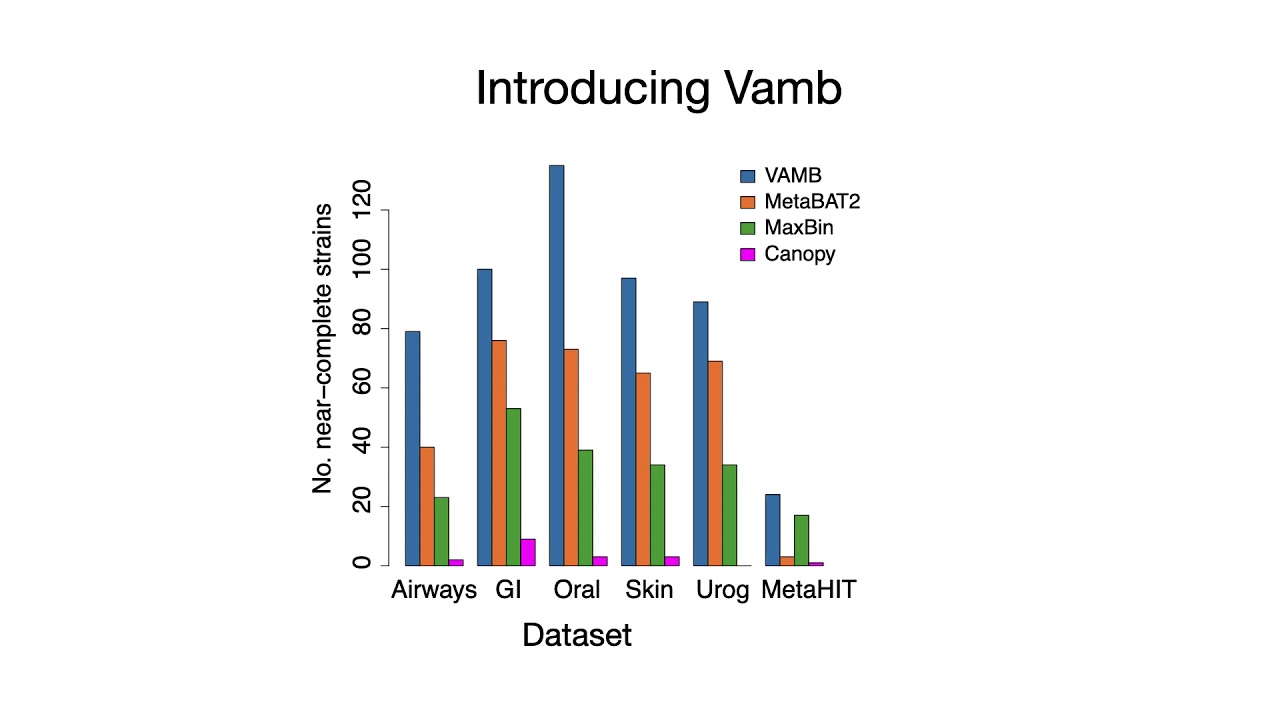
Despite recent advances in metagenomic binning, reconstruction of microbial species from metagenomics data, remains a challenging task. We have used recent advances in deep learning to develop Variational Autoencoders for Metagenomic Binning (VAMB), a program that uses deep variational autoencoders to encode sequence co-abundance and k-mer distribution information prior to clustering. We show that a variational autoencoder is able to integrate these two distinct data types without any prior knowledge of the datasets. VAMB outperforms existing state-of-the-art binners on contig datasets, reconstructing 29–98% more near complete draft genomes. We employed VAMB in a novel multi-split workflow, that enables assembly of 28–105% more strains compared to using VAMB with the commonly used single sample binning strategy. To demonstrate the scalability of our method, we bin a human gut microbiome dataset with 1,000 samples and reconstruct 45% more near-complete bins compared to state-of-the-art methods. Furthermore, we show that VAMB enables direct high-resolution taxonomic analysis of the generated genome clusters. Finally, we use this to show that different organisms have different geographical distribution patterns potentially important for design of probiotics. VAMB can be run on standard hardware and is freely available at [ Ссылка ].